Protein Post-translational Modification
Research in our group focuses on the chemistry and biology of protein post-translational modification. Post-translational modifications play an essential role in regulating protein function, with these modifications linked to multiple human diseases such as diabetes, cancer, cardiovascular disease, obesity, and substance abuse/addiction. The enzymes responsible for these chemical transformations face a daunting molecular recognition challenge within the cell – they need to efficiently identify and modify their substrates within a complex mixture of other proteins. Understanding the strategies employed by these enzymes will help identify novel substrates, while potentially illuminating new targets for therapeutic intervention and inhibitor development.
Drawing from chemistry, biochemistry, and molecular biology, our research program investigates enzymes involved in post-translational modification and the substrate proteins they act upon. We focus on proteins involved in protein lipidation pathways, such as acyltransferases, prenyltransferases, and related enzymes. These enzymes alter protein hydrophobicity through covalent modification, leading to changes in protein localization, protein-protein interactions, and biological activity. In each system, our laboratory investigates enzymatic strategies used to recognize substrates and catalyze lipidations, the effects of lipid modification(s) on protein function, and the biological impact of these modifications on cellular and organismal function.
Research projects
1. Ghrelin acylation by ghrelin O-acyltransferase
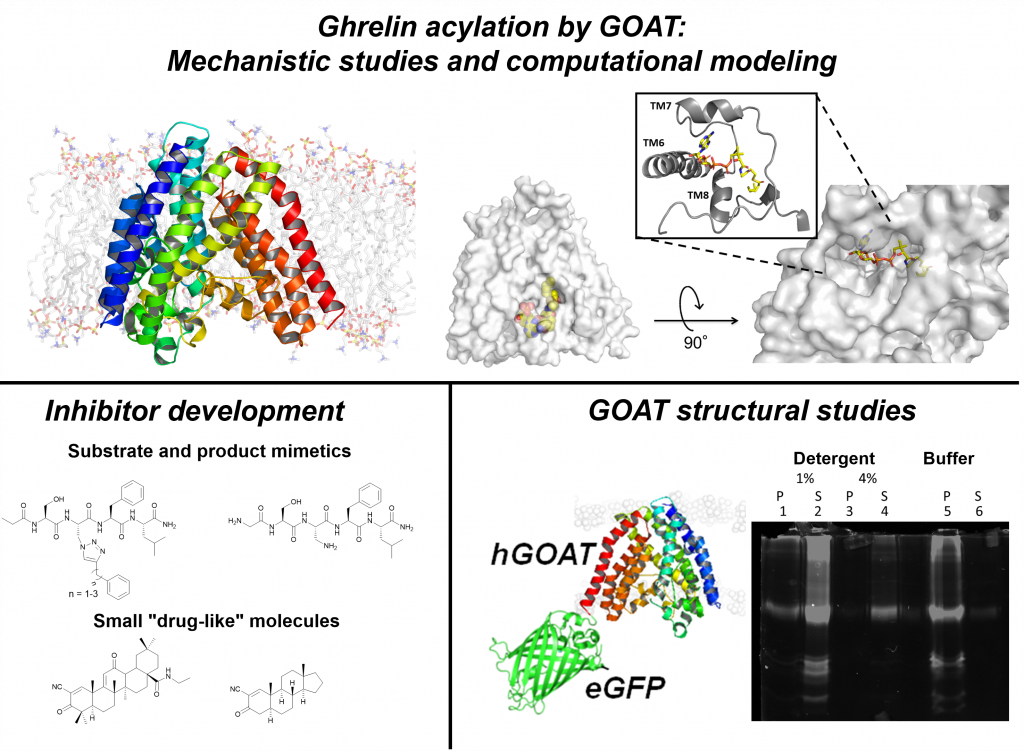
Secreted proteins play roles in controlling essential biological processes including cell growth, differentiation, and energy metabolism. Enzymes in the membrane-bound O-acyltransferase (MBOAT) family modify multiple secreted signaling proteins, such as ghrelin, Hedgehog, and Wnt. Discovery of ghrelin and its effects on appetite and glucose metabolism suggests that ghrelin-influenced pathways can provide an avenue for treatment of obesity and diabetes.
Ghrelin undergoes a unique serine O-octanoylation modification that is essential for binding to its receptor to activate signaling. Ghrelin O-acyltransferase (GOAT), the enzyme that catalyzes this modification, presents an attractive target point for controlling ghrelin signaling. We have shown ghrelin is a unique substrate for GOAT within the human proteome, and using computational modeling coupled with biochemical studies we have developed a structural model for GOAT. In our continuing studies of ghrelin acylation by GOAT, we are:
- Determining GOAT’s active site architecture and catalytic mechanism
- Purifying GOAT to enable experimental structural studies
- Applying library screening and rational design to create potent GOAT inhibitors
2. Exploring the localization, trafficking, and function of GOAT – expanding the ghrelin signaling pathway
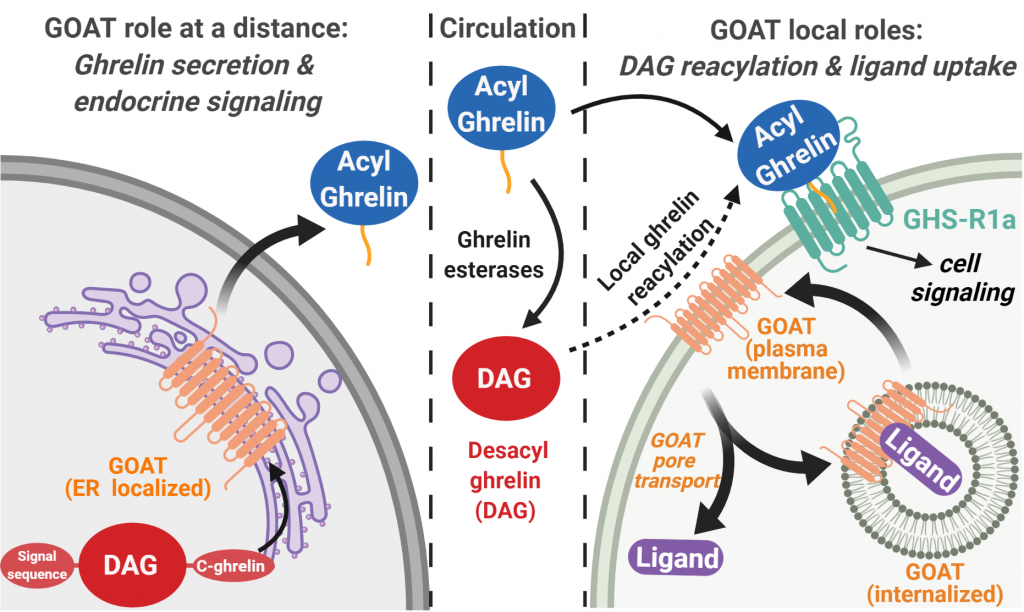
GOAT was originally identified to reside in the endoplasmic reticulum, where it catalyzes ghrelin octanoylation as part of the ghrelin processing and secretion pathway. However, recent work suggests GOAT could also reside on the cell’s plasma membrane where it would be exposed to the extracellular environment. On the cell membrane, GOAT could bind and re-acylate ghrelin as part of a local ghrelin signaling mechanism complementary to ghrelin’s well-established endocrine (long-range) signaling role within the body.
We are exploring the cellular localization and trafficking of GOAT using chemical biology, protein engineering, cell biology, and fluorescence imaging. Our work will establish a key step required for autocrine/paracrine ghrelin signaling involving local reacylation by GOAT, enable validation of GOAT itself as a disease biomarker and avenue for targeted treatments, and raise the possibility that other peptide hormones may exhibit similar complexity in their intercellular and organismal-level signaling pathways.
3. Defining the prenylated proteome: Investigating the biochemical foundation and biological impact of enzyme multispecificity in protein prenylation
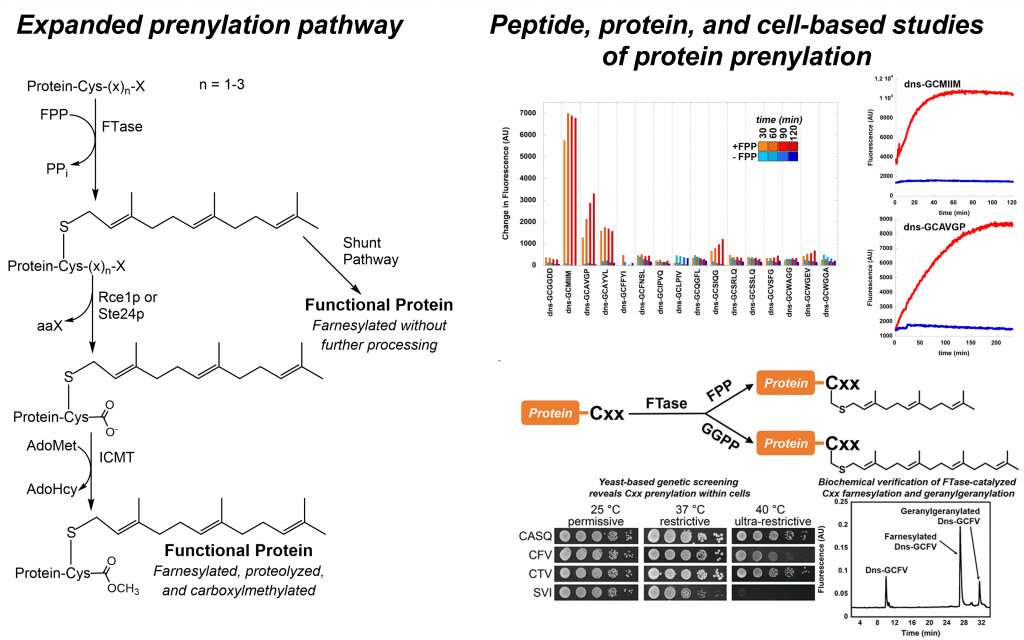
The prenyltransferases FTase and GGTase-I are examples of multispecific enzymes which must each modify a large number of protein substrates. The requirement to react with multiple substrates while maintaining selectivity against non-substrates presents a formidable molecular recognition challenge, one requiring both flexibility and fidelity. FTase and GGTase-I catalyze the modification of potentially several hundred cellular proteins, providing two related enzyme-substrate systems for exploring the biochemical basis for enzyme recognition of multiple target sequences. Our studies address the fundamental interactions that engender substrate selectivity in these multispecific enzymes and explore the potential extent of the prenylated proteome.
Using chemical probes and biochemical tools in both enzyme- and cell-based studies, we are defining the scope and biological impact of protein prenylation. While both FTase and GGTase-I recognize a C-terminal “CaaX” amino acid sequence in their protein substrates, we have shown FTase can also efficiently modify longer “C(x)3X” and shorter “Cxx” sequences. Intriguingly, many of these alternate length non-canonical substrates do not undergo post-prenylation processing steps and appear to impact protein function through mechanisms beyond membrane localization/association. Our studies provide a new avenue for probing the selectivity of prenylation pathway enzymes, determining the effects of prenylation pathway modifications on the cellular function of a protein, and expand the scope of protein prenylation for studies of protein structure and function.
